In silico simulation ofTerminal Restriction Fragment Length Polymorphism (T-RFLP)This script is an implementacion of in silico PCR-RFLP simulation tool. In order to undertand how this tool works, we will simulate the following experiment:  PCR
amplification
The
selected primers will amplify a 328 bp fragment from 16S ribosomal
RNA.
In E. coli genome there are 7 copies of the
target sequence, so 7 amplicons will be obtained. .
One primer (in red) is labelled, and the other one is not. In this simulation tool, both bands may be labelled. 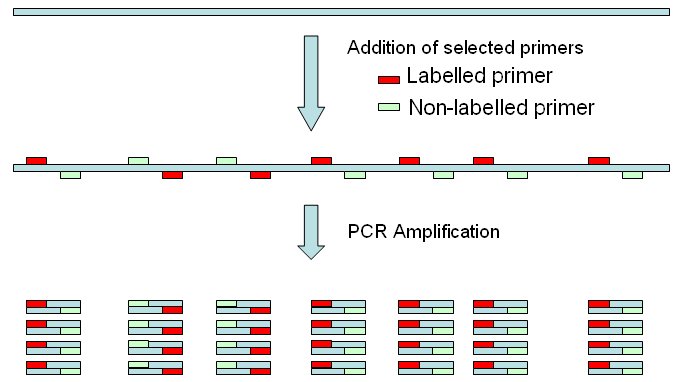 Endonuclease
restriction of amplicons and detection on fragments
One or two endonucleases may be used in the
experiment.
In this experiment only one endonuclease is used:
BspLI cuts at GGN'NCC and yields blunt
ends.
BspLI cleaves within one of the
amplified bands, yielding two fragments of 136 and 191bp.
All 328 bands and the 191 bp band are labelled. 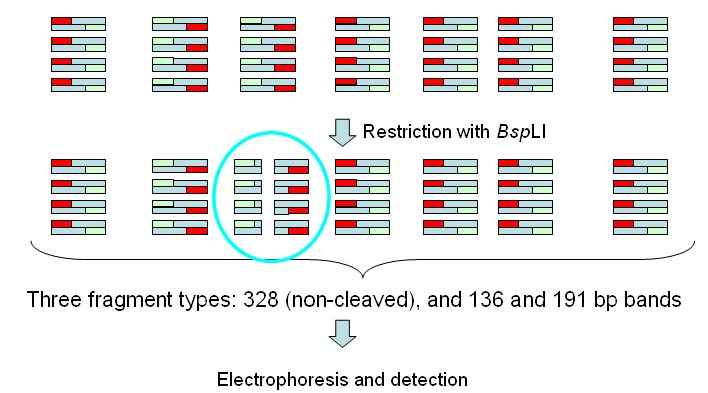 In the results page we will get a table like the one below: 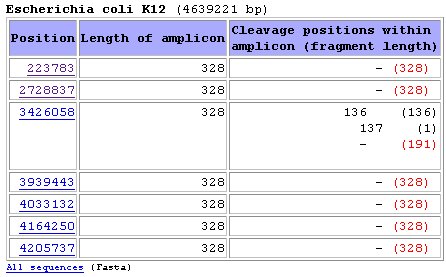 Position
means the exact nucleotide
within genome wheare the amplicon starts
Length of amplicon
is the length of the PCR product
Cleavage positions within
amplicon show the exact position where
restriction takes place. In this case, endonuclease BspLI cleaves the
third amplicon in the list at positions 136 and 137 (overlapping
cleavage sites are present). Althought fragments of sizes 136, 137, 191
and 192 will be yielded, the script will only computed bands of lengths
136 and 191 for simulation of the band patterns below.Fragment length is the length of each fragment. When the amplicon is not restricted the total length of the amplicon is shown Red fragment length: those are the labelled fragments (the ones containing the labelled primer) Simulation
of electrophoretic band pattern
Although detection of bands will often be performed by capillary electrophoresis, we will use simulated band patterns in a regular gel.
Only labelled fragments are shown. 
2003-2015@ University of the Basque Country. All
rights reserved.
|