In silico simulation of DNA fingerprintingThis tool will simulate the following general procedure:
Selection of genome When simulating this technique, in the first step we will be required to select a prokaryotic genera. After this selection, it is required to select a genome sequence, and other parameters for simulation of the experiment.. Design of probes: target sequence
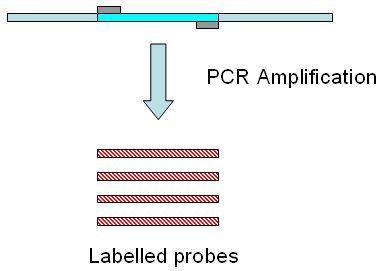
Target sequence is defined in this tool as the DNA sequence
we have obtained several copies from. The DNA copies will be labelled,
and they will be later used for hybridization againts a endonuclease digested
DNA fragments from the sample. These labelled copies are the probes.
This tool allows two procedures to define the target sequence:
Complete restriction digest of genome and electrophoresis of fragments After digestion, the sequence with capacity to hybridize with the probe (the target sequence) will be included in a unique fragment or in several ones. In the image below the target sequence (blue) is separated into 3 fragments of different sizes. 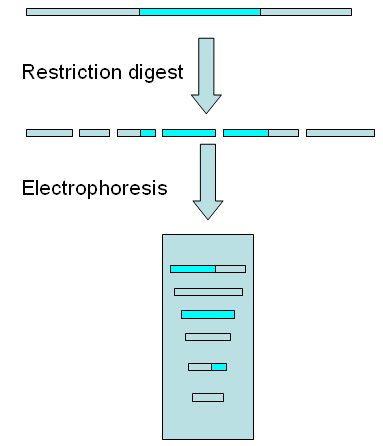
Hybridization with the probe and detection of bands Electophoretically separated DNA fragments are hybridized with previously developped probe (red). In this example, only 3 fragments will hybridize with the probe. Detection of bands will depend on labelling technique. 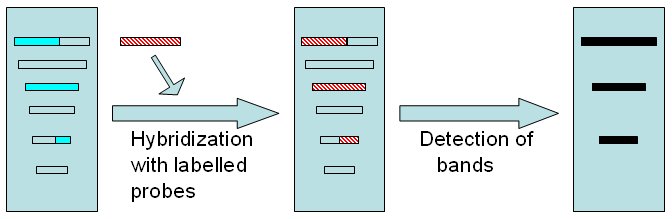
2005-2015@ University of the Basque Country. All rights
reserved.
|